Difference between revisions of "TnrA"
(→Categories containing this gene/protein) |
(→Reviews) |
||
| Line 149: | Line 149: | ||
=References= | =References= | ||
==Reviews== | ==Reviews== | ||
| − | <pubmed> 10231480 18086213 | + | <big>''Gunka K, Commichau FM'' </big> |
| + | <big>'''Control of glutamate homeostasis in ''Bacillus subtilis'':''' </big> | ||
| + | <big>'''a complex interplay between ammonium assimilation, glutamate biosynthesis and degradation. ''' </big> | ||
| + | <big>Mol Microbiol.: 2012, 85(2) 213-224. </big> | ||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/22625175 PubMed:22625175] | ||
| + | <pubmed> 10231480 18086213</pubmed> | ||
| + | |||
==The [[TnrA regulon]]== | ==The [[TnrA regulon]]== | ||
<pubmed>12823818,</pubmed> | <pubmed>12823818,</pubmed> | ||
Revision as of 17:44, 6 July 2012
- Description: transcriptional pleiotropic regulator invoved in global nitrogen regulation
| Gene name | tnrA |
| Synonyms | scgR |
| Essential | no |
| Product | transcription activator/ repressor |
| Function | regulation of nitrogen assimilation |
| Interactions involving this protein in SubtInteract: TnrA | |
| Metabolic function and regulation of this protein in SubtiPathways: Lipid synthesis, Nucleotides (regulation), Ile, Leu, Val, Ammonium/ glutamate, Central C-metabolism, Cell wall, Coenzyme A, Phosphorelay, Alternative nitrogen sources | |
| MW, pI | 12 kDa, 10.235 |
| Gene length, protein length | 330 bp, 110 aa |
| Immediate neighbours | mgtE, ykzB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 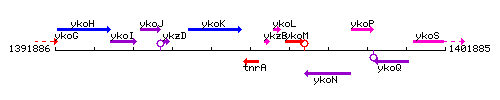
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed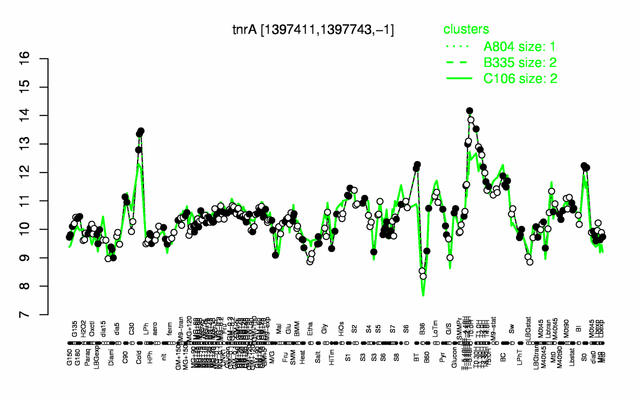
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids, glutamate metabolism, transcription factors and their control, regulators of core metabolism
This gene is a member of the following regulons
The TnrA regulon
The gene
Basic information
- Locus tag: BSU13310
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity: feedback-inhibited GlnA prevents TnrA from DNA binding
- Localization: membrane-associated via NrgA-NrgB under conditions of poor nitrogen supply PubMed
Database entries
- Structure:
- UniProt: Q45666
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: tnrA (according to DBTBS)
- Sigma factor:
- Regulation:
- Additional information:
Biological materials
- Mutant: GP252 (in frame deletion), available in the Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody: available in the Karl Forchhammer lab
Labs working on this gene/protein
Susan Fisher, Boston, USA homepage
Your additional remarks
References
Reviews
Gunka K, Commichau FM Control of glutamate homeostasis in Bacillus subtilis: a complex interplay between ammonium assimilation, glutamate biosynthesis and degradation. Mol Microbiol.: 2012, 85(2) 213-224. PubMed:22625175
The TnrA regulon
Control of TnrA activity by the trigger enzyme GlnA
Other original publications
Additional publications: PubMed