Difference between revisions of "LigA"
| Line 24: | Line 24: | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:ligA_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:ligA_context.gif]] | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
| + | |- | ||
| + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=ligA_721613_723619_1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:ligA_expression.png|500px]] | ||
|- | |- | ||
|} | |} | ||
__TOC__ | __TOC__ | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | <br/><br/><br/><br/> | ||
| + | |||
<br/><br/> | <br/><br/> | ||
Revision as of 15:30, 18 April 2012
- Description: DNA ligase (NAD-dependent)
| Gene name | ligA |
| Synonyms | lig, yerG |
| Essential | yes PubMed |
| Product | DNA ligase (NAD-dependent) |
| Function | replication |
| MW, pI | 74 kDa, 4.708 |
| Gene length, protein length | 2004 bp, 668 aa |
| Immediate neighbours | pcrA, yerH |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 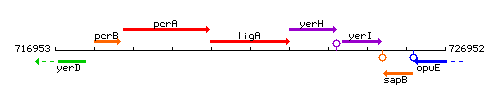
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed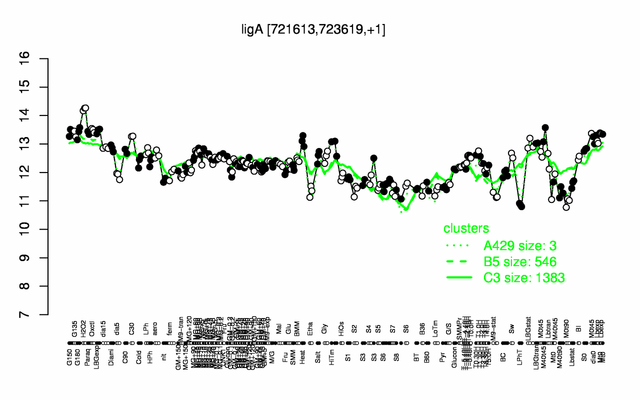
| |
Contents
Categories containing this gene/protein
DNA replication, essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU06620
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: NAD+ + (deoxyribonucleotide)(n) + (deoxyribonucleotide)(m) = AMP + nicotinamide nucleotide + (deoxyribonucleotide)(n+m) (according to Swiss-Prot)
- Protein family: BRCT domain (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure: 1B04 (adenylation domain, Geobacillus stearothermophilus)
- UniProt: O31498
- KEGG entry: [3]
- E.C. number: 6.5.1.2
Additional information
Expression and regulation
- Sigma factor:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Nora Au, Elke Kuester-Schoeck, Veena Mandava, Laura E Bothwell, Susan P Canny, Karen Chachu, Sierra A Colavito, Shakierah N Fuller, Eli S Groban, Laura A Hensley, Theresa C O'Brien, Amish Shah, Jessica T Tierney, Louise L Tomm, Thomas M O'Gara, Alexi I Goranov, Alan D Grossman, Charles M Lovett
Genetic composition of the Bacillus subtilis SOS system.
J Bacteriol: 2005, 187(22);7655-66
[PubMed:16267290]
[WorldCat.org]
[DOI]
(P p)
M A Petit, S D Ehrlich
The NAD-dependent ligase encoded by yerG is an essential gene of Bacillus subtilis.
Nucleic Acids Res: 2000, 28(23);4642-8
[PubMed:11095673]
[WorldCat.org]
[DOI]
(I p)
M A Petit, E Dervyn, M Rose, K D Entian, S McGovern, S D Ehrlich, C Bruand
PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication.
Mol Microbiol: 1998, 29(1);261-73
[PubMed:9701819]
[WorldCat.org]
[DOI]
(P p)