Difference between revisions of "Pgk"
(→Phenotypes of a mutant) |
|||
| Line 108: | Line 108: | ||
** ''[[cggR]]-[[gapA]]-[[pgk]]-[[tpiA]]-[[pgm]]-[[eno]]'' {{PubMed|11489127}} | ** ''[[cggR]]-[[gapA]]-[[pgk]]-[[tpiA]]-[[pgm]]-[[eno]]'' {{PubMed|11489127}} | ||
** ''[[pgk]]-[[tpiA]]-[[pgm]]-[[eno]]'' {{PubMed|11489127}} | ** ''[[pgk]]-[[tpiA]]-[[pgm]]-[[eno]]'' {{PubMed|11489127}} | ||
| + | |||
| + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=pgk_3480197_3481381_-1 pgk] {{PubMed|22383849}} | ||
* '''Sigma factor:''' | * '''Sigma factor:''' | ||
Revision as of 06:44, 17 April 2012
- Description: phosphoglycerate kinase, glycolytic/ gluconeogenic enzyme, universally conserved protein
| Gene name | pgk |
| Synonyms | |
| Essential | No |
| Product | phosphoglycerate kinase |
| Function | enzyme in glycolysis/ gluconeogenesis |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 42,0 kDa, 4.77 |
| Gene length, protein length | 1182 bp, 394 amino acids |
| Immediate neighbours | tpi, gapA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 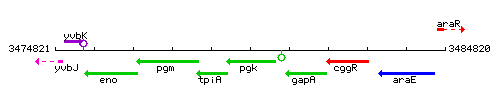
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
ATP synthesis, carbon core metabolism, phosphoproteins, universally conserved proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU33930
Phenotypes of a mutant
- no growth on LB medium, the mutant requires glucose and malate for growth
- essential according to Kobayashi et al. PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + 3-phospho-D-glycerate = ADP + 1,3-bisphosphoglycerate (according to Swiss-Prot)
- Protein family: phosphoglycerate kinase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information: Two Substrate Reversible Michaelis-Menten PubMed
- Domains:
- nucleotide binding domain (ATP) (350–353)
- 2x substrate binding domain (21–23), (59–62)
- Cofactor(s): Mg2+ or Mn2+ PubMed
- Effectors of protein activity:
- Inhibited by Co2+, NDP and NMP PubMed
- Localization: cytoplasm (according to Swiss-Prot), Cytoplasm (Homogeneous) PubMed
Database entries
- Structure: 1PHP (from Geobacillus stearothermophilus)
- UniProt: P40924
- KEGG entry: [3]
- E.C. number: 2.7.2.3
Additional information
- extensive information on the structure and enzymatic properties of Pgk can be found at Proteopedia
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Expression vector:
- pGP1102 (N-terminal His-tag, in pWH844), available in Stülke lab
- pGP95 (N-terminal Strep-tag, in pGP172), available in Stülke lab
- pGP91 (N-terminal Strep-tag, for SPINE, expression in B. subtilis, in pGP380), available in Stülke lab
- pGP1513 (expression in B. subtilis, in pBQ200), available in Stülke lab
- lacZ fusion: pGP514 (in pAC6), a series of promoter deletion variants is also available in pAC6, available in Stülke lab
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Christine Eymann, Dörte Becher, Jörg Bernhardt, Katrin Gronau, Anja Klutzny, Michael Hecker
Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis.
Proteomics: 2007, 7(19);3509-26
[PubMed:17726680]
[WorldCat.org]
[DOI]
(P p)
Laurent Jannière, Danielle Canceill, Catherine Suski, Sophie Kanga, Bérengère Dalmais, Roxane Lestini, Anne-Françoise Monnier, Jérôme Chapuis, Alexander Bolotin, Marina Titok, Emmanuelle Le Chatelier, S Dusko Ehrlich
Genetic evidence for a link between glycolysis and DNA replication.
PLoS One: 2007, 2(5);e447
[PubMed:17505547]
[WorldCat.org]
[DOI]
(I e)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Jean-Christophe Meile, Ling Juan Wu, S Dusko Ehrlich, Jeff Errington, Philippe Noirot
Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory.
Proteomics: 2006, 6(7);2135-46
[PubMed:16479537]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
K Kobayashi, S D Ehrlich, A Albertini, G Amati, K K Andersen, M Arnaud, K Asai, S Ashikaga, S Aymerich, P Bessieres, F Boland, S C Brignell, S Bron, K Bunai, J Chapuis, L C Christiansen, A Danchin, M Débarbouille, E Dervyn, E Deuerling, K Devine, S K Devine, O Dreesen, J Errington, S Fillinger, S J Foster, Y Fujita, A Galizzi, R Gardan, C Eschevins, T Fukushima, K Haga, C R Harwood, M Hecker, D Hosoya, M F Hullo, H Kakeshita, D Karamata, Y Kasahara, F Kawamura, K Koga, P Koski, R Kuwana, D Imamura, M Ishimaru, S Ishikawa, I Ishio, D Le Coq, A Masson, C Mauël, R Meima, R P Mellado, A Moir, S Moriya, E Nagakawa, H Nanamiya, S Nakai, P Nygaard, M Ogura, T Ohanan, M O'Reilly, M O'Rourke, Z Pragai, H M Pooley, G Rapoport, J P Rawlins, L A Rivas, C Rivolta, A Sadaie, Y Sadaie, M Sarvas, T Sato, H H Saxild, E Scanlan, W Schumann, J F M L Seegers, J Sekiguchi, A Sekowska, S J Séror, M Simon, P Stragier, R Studer, H Takamatsu, T Tanaka, M Takeuchi, H B Thomaides, V Vagner, J M van Dijl, K Watabe, A Wipat, H Yamamoto, M Yamamoto, Y Yamamoto, K Yamane, K Yata, K Yoshida, H Yoshikawa, U Zuber, N Ogasawara
Essential Bacillus subtilis genes.
Proc Natl Acad Sci U S A: 2003, 100(8);4678-83
[PubMed:12682299]
[WorldCat.org]
[DOI]
(P p)
H Ludwig, G Homuth, M Schmalisch, F M Dyka, M Hecker, J Stülke
Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon.
Mol Microbiol: 2001, 41(2);409-22
[PubMed:11489127]
[WorldCat.org]
[DOI]
(P p)
K Suzuki, K Imahori
Phosphoglycerate kinase from Bacillus stearothermophilus.
Methods Enzymol: 1982, 90 Pt E;126-30
[PubMed:7154941]
[WorldCat.org]
[DOI]
(P p)