Difference between revisions of "YesT"
m (Reverted edits by 134.76.70.252 (talk) to last revision by Jstuelk) |
|||
| Line 98: | Line 98: | ||
* '''Operon:''' ''[[yesS]]-[[yesT]]-[[yesU]]-[[yesV]]-[[yesW]]-[[yesX]]-[[yesY]]-[[yesZ]]'' {{PubMed|19651770}} | * '''Operon:''' ''[[yesS]]-[[yesT]]-[[yesU]]-[[yesV]]-[[yesW]]-[[yesX]]-[[yesY]]-[[yesZ]]'' {{PubMed|19651770}} | ||
| − | * '''[ | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=rhgT_768137_768835_1 yesT] {{PubMed|22383849}} |
| + | |||
| + | * '''Sigma factor:''' | ||
* '''Regulation:''' induced by pectin ([[YesS]]) {{PubMed|19651770}} | * '''Regulation:''' induced by pectin ([[YesS]]) {{PubMed|19651770}} | ||
| Line 104: | Line 106: | ||
* '''Regulatory mechanism:''' [[YesS]]: transcriptional activation {{PubMed|19651770}} | * '''Regulatory mechanism:''' [[YesS]]: transcriptional activation {{PubMed|19651770}} | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
Revision as of 15:47, 12 April 2012
- Description: rhamnogalacturonan acetylesterase
| Gene name | yesT |
| Synonyms | |
| Essential | no |
| Product | rhamnogalacturonan esterase |
| Function | pectin utilization |
| MW, pI | 25 kDa, 5.505 |
| Gene length, protein length | 696 bp, 232 aa |
| Immediate neighbours | yesS, yesU |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 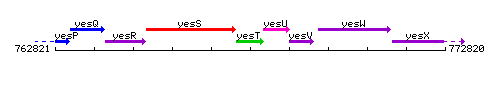
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU07020
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: 'GDSL' lipolytic enzyme family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O31523
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References