Difference between revisions of "SigB"
(→Identification of the SigB regulon) |
(→Other publications) |
||
| Line 157: | Line 157: | ||
<big>Science. 2011 334:366-369. </big> | <big>Science. 2011 334:366-369. </big> | ||
[http://www.ncbi.nlm.nih.gov/pubmed/21979936 PubMed:21979936] | [http://www.ncbi.nlm.nih.gov/pubmed/21979936 PubMed:21979936] | ||
| − | <pubmed> 8655572 | + | <pubmed> 22210769, 8655572, 3123466, 10369900,3112122, 8468294,8458834, 13129942, 6784117, 12867438, 3100810, 8253681, 12486038, 8764398,3027048, 15342585,17575448, 17586624,,3016731,11902719,14651641,10503549,15205443,6405278, 10383961, 15805528 6790515 116131, 39767581 19948797 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 13:30, 3 January 2012
- Description: RNA polymerase sigma factor SigB
| Gene name | sigB |
| Synonyms | rpoF |
| Essential | no |
| Product | RNA polymerase sigma factor SigB |
| Function | general stress response |
| Interactions involving this protein in SubtInteract: SigB | |
| Metabolic function and regulation of this protein in SubtiPathways: Stress, Murein recycling | |
| MW, pI | 29 kDa, 5.418 |
| Gene length, protein length | 792 bp, 264 aa |
| Immediate neighbours | rsbW, rsbX |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 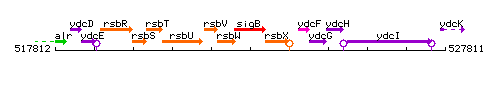
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
transcription, sigma factors and their control, general stress proteins (controlled by SigB)
This gene is a member of the following regulons
The SigB regulon
The gene
Basic information
- Locus tag: BSU04730
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: SigB subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P06574
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant: QB5344 (cat), available in the Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Bill Haldenwang, San Antonio, USA
- Chet Price, Davis, USA homepage
Your additional remarks
References
Reviews
Control of SigB activity by protein-protein interactions
Identification of the SigB regulon
Priyanka Nannapaneni, Falk Hertwig, Maren Depke, Michael Hecker, Ulrike Mäder, Uwe Völker, Leif Steil, Sacha A F T van Hijum
Defining the structure of the general stress regulon of Bacillus subtilis using targeted microarray analysis and random forest classification.
Microbiology (Reading): 2012, 158(Pt 3);696-707
[PubMed:22174379]
[WorldCat.org]
[DOI]
(I p)
J D Helmann, M F Wu, P A Kobel, F J Gamo, M Wilson, M M Morshedi, M Navre, C Paddon
Global transcriptional response of Bacillus subtilis to heat shock.
J Bacteriol: 2001, 183(24);7318-28
[PubMed:11717291]
[WorldCat.org]
[DOI]
(P p)
A Petersohn, M Brigulla, S Haas, J D Hoheisel, U Völker, M Hecker
Global analysis of the general stress response of Bacillus subtilis.
J Bacteriol: 2001, 183(19);5617-31
[PubMed:11544224]
[WorldCat.org]
[DOI]
(P p)
C W Price, P Fawcett, H Cérémonie, N Su, C K Murphy, P Youngman
Genome-wide analysis of the general stress response in Bacillus subtilis.
Mol Microbiol: 2001, 41(4);757-74
[PubMed:11532142]
[WorldCat.org]
[DOI]
(P p)
A Petersohn, J Bernhardt, U Gerth, D Höper, T Koburger, U Völker, M Hecker
Identification of sigma(B)-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization.
J Bacteriol: 1999, 181(18);5718-24
[PubMed:10482513]
[WorldCat.org]
[DOI]
(P p)
Anja Petersohn, Haike Antelmann, Ulf Gerth, Michael Hecker
Identification and transcriptional analysis of new members of the sigmaB regulon in Bacillus subtilis.
Microbiology (Reading): 1999, 145 ( Pt 4);869-880
[PubMed:10220166]
[WorldCat.org]
[DOI]
(P p)
Other publications
Locke JC, Young JW, Fontes M, Hernández Jiménez MJ, Elowitz MB Stochastic pulse regulation in bacterial stress response. Science. 2011 334:366-369. PubMed:21979936