Difference between revisions of "PutB"
(→Extended information on the protein) |
|||
| Line 1: | Line 1: | ||
| − | * '''Description:''' proline | + | * '''Description:''' proline dehydrogenase (L-proline, NAD) <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || proline | + | |style="background:#ABCDEF;" align="center"| '''Product''' || proline dehydrogenase (L-proline, NAD) |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || proline utilization | |style="background:#ABCDEF;" align="center"|'''Function''' || proline utilization | ||
| Line 57: | Line 57: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| Line 95: | Line 93: | ||
* '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU03200] | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU03200] | ||
| − | * '''E.C. number:''' | + | * '''E.C. number:''' [http://www.expasy.org/enzyme/1.5.99.8 1.5.99.8] |
=== Additional information=== | === Additional information=== | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''[[putB]]-[[putC]]-[[putP]]'' {{PubMed|21840319}} | + | * '''Operon:''' ''[[putB]]-[[putC]]-[[putP]]'' {{PubMed|21840319, 22139509}} |
| − | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|21840319,21964733}} | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|21840319,21964733,22139509}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| − | ** induced in the presence of proline ([[PutR]]) {{PubMed|21840319}} | + | ** induced in the presence of proline ([[PutR]]) {{PubMed|21840319,22139509}} |
** expressed under conditions that trigger sporulation ([[Spo0A]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/14651647 PubMed] | ** expressed under conditions that trigger sporulation ([[Spo0A]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/14651647 PubMed] | ||
** repressed during growth in the presence of branched chain amino acids ([[CodY]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/12618455 PubMed] | ** repressed during growth in the presence of branched chain amino acids ([[CodY]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/12618455 PubMed] | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | ** [[PutR]]: transcription activation {{PubMed|21840319,21964733}} | + | ** [[PutR]]: transcription activation {{PubMed|21840319,21964733,22139509}} |
** [[Spo0A]]: transcription activation [http://www.ncbi.nlm.nih.gov/sites/entrez/14651647 PubMed] | ** [[Spo0A]]: transcription activation [http://www.ncbi.nlm.nih.gov/sites/entrez/14651647 PubMed] | ||
** [[CodY]]: transcription repression, displacement of [[PutR]] {{PubMed|21840319}} | ** [[CodY]]: transcription repression, displacement of [[PutR]] {{PubMed|21840319}} | ||
| Line 137: | Line 135: | ||
=References= | =References= | ||
'''Additional publications:''' {{PubMed|21964733}} | '''Additional publications:''' {{PubMed|21964733}} | ||
| − | <pubmed>14976255,21840319,14651647,12618455, </pubmed> | + | <pubmed>14976255,21840319,14651647,12618455, 22139509 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 12:21, 9 December 2011
- Description: proline dehydrogenase (L-proline, NAD)
| Gene name | putB |
| Synonyms | ycgM |
| Essential | no |
| Product | proline dehydrogenase (L-proline, NAD) |
| Function | proline utilization |
| Metabolic function and regulation of this protein in SubtiPathways: Ammonium/ glutamate | |
| MW, pI | 34 kDa, 7.291 |
| Gene length, protein length | 909 bp, 303 aa |
| Immediate neighbours | ycgL, putC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 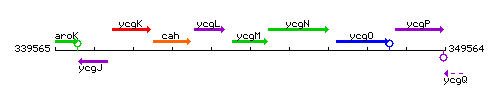
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
CodY regulon, PutR regulon, Spo0A regulon
The gene
Basic information
- Locus tag: BSU03200
Phenotypes of a mutant
- no growth with proline as single source of carbon or nitrogen PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: L-proline + acceptor = (S)-1-pyrroline-5-carboxylate + reduced acceptor (according to Swiss-Prot)
- Protein family: proline dehydrogenase family (according to Swiss-Prot)
- Paralogous protein(s): FadM
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P94390
- KEGG entry: [3]
- E.C. number: 1.5.99.8
Additional information
Expression and regulation
- Operon: putB-putC-putP 22139509 PubMed
- Regulation:
- Regulatory mechanism:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Additional publications: PubMed