Difference between revisions of "PyrG"
| Line 1: | Line 1: | ||
| − | + | * '''Description:''' CTP synthase (NH3, glutamine) <br/><br/> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | * '''Description:''' CTP synthase (NH3, glutamine) | ||
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 22: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || pyrimidine biosynthesis | |style="background:#ABCDEF;" align="center"|'''Function''' || pyrimidine biosynthesis | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/pyrimidines_metabolism.html Pyrimidines], [http://subtiwiki.uni-goettingen.de/pathways/gene_regulation_nucleotides.html Nucleotides (regulation)]''' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 59 kDa, 5.16 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 59 kDa, 5.16 | ||
| Line 30: | Line 22: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ywjG]]'', ''[[rpoE]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ywjG]]'', ''[[rpoE]]'' | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+& | + | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB15743]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' |
|- | |- | ||
| − | |colspan="2" | '''Genetic context''' | + | |colspan="2" | '''Genetic context''' <br/> [[Image:pyrG_context.gif]] |
| − | + | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | |
|- | |- | ||
|} | |} | ||
| Line 39: | Line 31: | ||
__TOC__ | __TOC__ | ||
| − | + | <br/><br/><br/><br/><br/><br/> | |
=The gene= | =The gene= | ||
| Line 64: | Line 56: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' ATP + UTP + NH | + | * '''Catalyzed reaction/ biological activity:''' ATP + UTP + NH<sub>3</sub> = ADP + phosphate + CTP (according to Swiss-Prot) |
* '''Protein family:''' CTP synthase family (according to Swiss-Prot) | * '''Protein family:''' CTP synthase family (according to Swiss-Prot) | ||
| Line 132: | Line 124: | ||
=References= | =References= | ||
| − | + | <pubmed>8206849,17302819,15252202,11544212,1709162,12426364,17158658,2457578 </pubmed> | |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 05:13, 24 November 2010
- Description: CTP synthase (NH3, glutamine)
| Gene name | pyrG |
| Synonyms | ctrA |
| Essential | yes PubMed |
| Product | CTP synthase (NH3, glutamine) |
| Function | pyrimidine biosynthesis |
| Metabolic function and regulation of this protein in SubtiPathways: Pyrimidines, Nucleotides (regulation) | |
| MW, pI | 59 kDa, 5.16 |
| Gene length, protein length | 1605 bp, 535 aa |
| Immediate neighbours | ywjG, rpoE |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 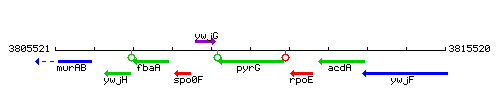
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU37150
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + UTP + NH3 = ADP + phosphate + CTP (according to Swiss-Prot)
- Protein family: CTP synthase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- UniProt: P13242
- KEGG entry: [3]
- E.C. number: 6.3.4.2
Additional information
Expression and regulation
- Operon: pyrG PubMed
- Regulation:
- repressed by cytidine nucleotides (RNA switch) PubMed
- Regulatory mechanism:
- RNA switch: transcription (termination in the presence of cytidine nucleotides) PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Irene E Jensen-MacAllister, Qi Meng, Robert L Switzer
Regulation of pyrG expression in Bacillus subtilis: CTP-regulated antitermination and reiterative transcription with pyrG templates in vitro.
Mol Microbiol: 2007, 63(5);1440-52
[PubMed:17302819]
[WorldCat.org]
[DOI]
(P p)
Alexander K W Elsholz, Casper Møller Jørgensen, Robert L Switzer
The number of G residues in the Bacillus subtilis pyrG initially transcribed region governs reiterative transcription-mediated regulation.
J Bacteriol: 2007, 189(5);2176-80
[PubMed:17158658]
[WorldCat.org]
[DOI]
(P p)
Qi Meng, Charles L Turnbough, Robert L Switzer
Attenuation control of pyrG expression in Bacillus subtilis is mediated by CTP-sensitive reiterative transcription.
Proc Natl Acad Sci U S A: 2004, 101(30);10943-8
[PubMed:15252202]
[WorldCat.org]
[DOI]
(P p)
Qi Meng, Robert L Switzer
cis-acting sequences of Bacillus subtilis pyrG mRNA essential for regulation by antitermination.
J Bacteriol: 2002, 184(23);6734-8
[PubMed:12426364]
[WorldCat.org]
[DOI]
(P p)
Q Meng, R L Switzer
Regulation of transcription of the Bacillus subtilis pyrG gene, encoding cytidine triphosphate synthetase.
J Bacteriol: 2001, 183(19);5513-22
[PubMed:11544212]
[WorldCat.org]
[DOI]
(P p)
R J Turner, Y Lu, R L Switzer
Regulation of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster by an autogenous transcriptional attenuation mechanism.
J Bacteriol: 1994, 176(12);3708-22
[PubMed:8206849]
[WorldCat.org]
[DOI]
(P p)
C L Quinn, B T Stephenson, R L Switzer
Functional organization and nucleotide sequence of the Bacillus subtilis pyrimidine biosynthetic operon.
J Biol Chem: 1991, 266(14);9113-27
[PubMed:1709162]
[WorldCat.org]
(P p)
K Trach, J W Chapman, P Piggot, D LeCoq, J A Hoch
Complete sequence and transcriptional analysis of the spo0F region of the Bacillus subtilis chromosome.
J Bacteriol: 1988, 170(9);4194-208
[PubMed:2457578]
[WorldCat.org]
[DOI]
(P p)