Difference between revisions of "OhrR"
(→Reviews) |
|||
| Line 64: | Line 64: | ||
* '''Domains:''' | * '''Domains:''' | ||
| − | * '''Modification:''' | + | * '''Modification:''' senses organic peroxides, released from DNA upon oxidation of an active site cysteine |
* '''Cofactor(s):''' | * '''Cofactor(s):''' | ||
| Line 76: | Line 76: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1Z91 1Z91] (reduced form), [http://www.rcsb.org/pdb/explore.do?structureId=1Z9C 1Z9C] (complex with ohrA promoter) | + | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1Z91 1Z91] (reduced form), [http://www.rcsb.org/pdb/explore.do?structureId=1Z9C 1Z9C] (complex with ''[[ohrA]]'' promoter) |
* '''UniProt:''' [http://www.uniprot.org/uniprot/O34777 O34777] | * '''UniProt:''' [http://www.uniprot.org/uniprot/O34777 O34777] | ||
| Line 90: | Line 90: | ||
* '''Operon:''' ''ohrR'' {{PubMed|9696771}} | * '''Operon:''' ''ohrR'' {{PubMed|9696771}} | ||
| − | * '''[[Sigma factor]]:''' | + | * '''[[Sigma factor]]:''' [[SigA]] |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 122: | Line 122: | ||
==Reviews== | ==Reviews== | ||
<pubmed>19575568 20094649 </pubmed> | <pubmed>19575568 20094649 </pubmed> | ||
| − | |||
==Original Publications== | ==Original Publications== | ||
<pubmed>16209951,18487332,17660290,17502599,12486061,11418552,18586944,11983871,18363800,19129220 9696771 </pubmed> | <pubmed>16209951,18487332,17660290,17502599,12486061,11418552,18586944,11983871,18363800,19129220 9696771 </pubmed> | ||
| − | |||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 18:21, 12 June 2010
- Description: transcription repressor of the ohrA gene
| Gene name | ohrR |
| Synonyms | ykmA |
| Essential | no |
| Product | transcription repressor (MarR family) |
| Function | regulation of ohrA expression in response to organic peroxides |
| MW, pI | 16 kDa, 6.364 |
| Gene length, protein length | 441 bp, 147 aa |
| Immediate neighbours | ohrA, ohrB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 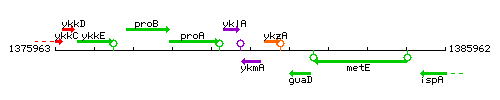
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU13150
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: senses organic peroxides, released from DNA upon oxidation of an active site cysteine
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- UniProt: O34777
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: ohrR PubMed
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
John Helmann, Cornell University, USA Homepage
Richard Brennan, Houston, Texas, USA Homepage
Your additional remarks
References
Reviews
Original Publications