Difference between revisions of "AraM"
(→Expression and regulation) |
|||
| Line 89: | Line 89: | ||
=Expression and regulation= | =Expression and regulation= | ||
| + | * '''Operon:''' ''[[araA]]-[[araB]]-[[araD]]-[[araL]]-[[araM]]-[[araN]]-[[araP]]-[[araQ]]-[[abfA]]'' {{PubMed|9084180}} | ||
| − | * ''' | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|9084180}} |
| − | * '''[[ | + | * '''Regulation:''' |
| + | ** repressed by glucose ([[CcpA]]) {{PubMed|12949161}} | ||
| + | ** induced by arabinose ([[AraR]]) [http://www.ncbi.nlm.nih.gov/sites/entrez/10417639 PubMed] | ||
| − | * ''' | + | * '''Regulatory mechanism:''' |
| − | ** | + | ** [[CcpA]]: transcription repression {{PubMed|12949161}} |
| − | |||
| − | |||
** [[AraR]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/10417639 PubMed] | ** [[AraR]]: transcription repression [http://www.ncbi.nlm.nih.gov/sites/entrez/10417639 PubMed] | ||
Revision as of 20:27, 16 December 2009
- Description: glycerol-1-phosphate dehydrogenase, L-arabinose operon
| Gene name | araM |
| Synonyms | yseB |
| Essential | no |
| Product | glycerol-1-phosphate dehydrogenase |
| Function | biosynthesis of phosphoglycerolipids |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 42 kDa, 5.628 |
| Gene length, protein length | 1182 bp, 394 aa |
| Immediate neighbours | araN, araL |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 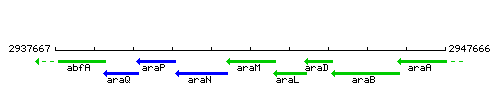
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU28760
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: sn-glycerol-1-phosphate + NAD(P)+ = glycerone phosphate + NAD(P)H (according to Swiss-Prot)
- Protein family: glycerol-1-phosphate dehydrogenase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P94527
- KEGG entry: [3]
- E.C. number: 1.1.1.261
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Harald Guldan, Reinhard Sterner, Patrick Babinger
Identification and characterization of a bacterial glycerol-1-phosphate dehydrogenase: Ni(2+)-dependent AraM from Bacillus subtilis.
Biochemistry: 2008, 47(28);7376-84
[PubMed:18558723]
[WorldCat.org]
[DOI]
(I p)
José Manuel Inácio, Carla Costa, Isabel de Sá-Nogueira
Distinct molecular mechanisms involved in carbon catabolite repression of the arabinose regulon in Bacillus subtilis.
Microbiology (Reading): 2003, 149(Pt 9);2345-2355
[PubMed:12949161]
[WorldCat.org]
[DOI]
(P p)
L J Mota, P Tavares, I Sá-Nogueira
Mode of action of AraR, the key regulator of L-arabinose metabolism in Bacillus subtilis.
Mol Microbiol: 1999, 33(3);476-89
[PubMed:10417639]
[WorldCat.org]
[DOI]
(P p)
Isabel Sa-Nogueira, Teresa V Nogueira, Snia Soares, Hermnia de Lencastre
The Bacillus subtilis L-arabinose (ara) operon: nucleotide sequence, genetic organization and expression.
Microbiology (Reading): 1997, 143 ( Pt 3);957-969
[PubMed:9084180]
[WorldCat.org]
[DOI]
(P p)