Difference between revisions of "SigB"
(→References) |
(→References) |
||
| Line 130: | Line 130: | ||
=References= | =References= | ||
| − | <pubmed> 19575568 8655572, | + | ==Reviews== |
| + | <pubmed> 19575568, 18035607, 39767581 </pubmed> | ||
| + | |||
| + | <pubmed> 8655572,, 3123466,8682769, 11717291,8460143,10369900,3112122,8468294,8458834, 13129942, 6784117, 12867438,3100810,8253681,12270815,12486038, 8764398,3027048,7601843,15342585,17575448, 15342582,10482513, 8002610,11591687,10220166,17586624,,3016731,11902719,14651641,10503549,15205443,6405278, 10383961,15312768, 17498739,11377871, 15805528 10482513 11544224 11532142 6790515 116131, </pubmed> | ||
Revision as of 10:26, 7 July 2009
- Description: RNA polymerase sigma factor SigB
| Gene name | sigB |
| Synonyms | rpoF |
| Essential | no |
| Product | RNA polymerase sigma factor SigB |
| Function | general stress response |
| Metabolic function and regulation of this protein in SubtiPathways: Stress | |
| MW, pI | 29 kDa, 5.418 |
| Gene length, protein length | 792 bp, 264 aa |
| Immediate neighbours | rsbW, rsbX |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 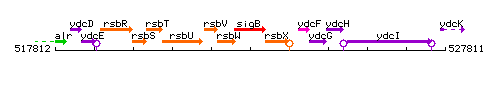
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU04730
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: SigB subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Genes/ operons controlled by SigB
csbB, ctc, gsiB, katE, lexA,licR, sfA, xpf, yaaH, ydeC, ytzE, yvaN, ywhH,
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure:
- Swiss prot entry: P06574
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Bill Haldenwang, San Antonio, USA
- Chet Price, Davis, USA homepage
Your additional remarks
References
Reviews