Difference between revisions of "RnpA"
| Line 82: | Line 82: | ||
* '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P25814 P25814] | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P25814 P25814] | ||
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU41050] |
* '''E.C. number:''' [http://www.expasy.org/enzyme/3.1.26.5 3.1.26.5] | * '''E.C. number:''' [http://www.expasy.org/enzyme/3.1.26.5 3.1.26.5] | ||
Revision as of 04:12, 25 June 2009
- Description: protein component of ribonuclease P
| Gene name | rnpA |
| Synonyms | |
| Essential | yes PubMed |
| Product | protein component of RNase P (substrate specificity) |
| Function | cleavage of precursor sequences from the 5' ends of pre-tRNAs |
| MW, pI | 13 kDa, 10.804 |
| Gene length, protein length | 348 bp, 116 aa |
| Immediate neighbours | spoIIIJ, rpmH |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 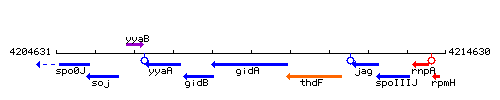
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU41050
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Endonucleolytic cleavage of RNA, removing 5'-extranucleotides from tRNA precursor (according to Swiss-Prot)
- Protein family: rnpA family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Swiss prot entry: P25814
- KEGG entry: [2]
- E.C. number: 3.1.26.5
Additional information
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Roland Hartmann, Marburg University, Germany homepage
Your additional remarks
References
Markus Gösringer, Roland K Hartmann
Function of heterologous and truncated RNase P proteins in Bacillus subtilis.
Mol Microbiol: 2007, 66(3);801-13
[PubMed:17919279]
[WorldCat.org]
[DOI]
(P p)
Barbara Wegscheid, Roland K Hartmann
In vivo and in vitro investigation of bacterial type B RNase P interaction with tRNA 3'-CCA.
Nucleic Acids Res: 2007, 35(6);2060-73
[PubMed:17355991]
[WorldCat.org]
[DOI]
(I p)
Markus Gössringer, Rosel Kretschmer-Kazemi Far, Roland K Hartmann
Analysis of RNase P protein (rnpA) expression in Bacillus subtilis utilizing strains with suppressible rnpA expression.
J Bacteriol: 2006, 188(19);6816-23
[PubMed:16980484]
[WorldCat.org]
[DOI]
(P p)
Christoph Rox, Ralph Feltens, Thomas Pfeiffer, Roland K Hartmann
Potential contact sites between the protein and RNA subunit in the Bacillus subtilis RNase P holoenzyme.
J Mol Biol: 2002, 315(4);551-60
[PubMed:11812129]
[WorldCat.org]
[DOI]
(P p)
A Hansen, T Pfeiffer, T Zuleeg, S Limmer, J Ciesiolka, R Feltens, R K Hartmann
Exploring the minimal substrate requirements for trans-cleavage by RNase P holoenzymes from Escherichia coli and Bacillus subtilis.
Mol Microbiol: 2001, 41(1);131-43
[PubMed:11454206]
[WorldCat.org]
[DOI]
(P p)
J M Warnecke, R Held, S Busch, R K Hartmann
Role of metal ions in the hydrolysis reaction catalyzed by RNase P RNA from Bacillus subtilis.
J Mol Biol: 1999, 290(2);433-45
[PubMed:10390342]
[WorldCat.org]
[DOI]
(P p)
T Stams, S Niranjanakumari, C A Fierke, D W Christianson
Ribonuclease P protein structure: evolutionary origins in the translational apparatus.
Science: 1998, 280(5364);752-5
[PubMed:9563955]
[WorldCat.org]
[DOI]
(P p)